lentil.smear#
- lentil.smear(img, distance, angle=None, pixelscale=1, oversample=1)[source]#
Apply image smear via convolution.
- Parameters:
img (array_like) – Input image
distance (float) –
Linear smear distance
If
pixelscale = 1,distanceshould give smear in terms of fractional pixels. Ifpixelscaleis the physical dimension of one pixel,distanceshould give smear in terms of the same units.angle (float, optional) – Smear direction in degrees measured clockwise from the x-axis. If
None(default), a new direction is randomly chosen every timeSmearis called.pixelscale (float, optional) – Pixel size. If
pixelscale = 1(default), thescaleparameter provided when Smear is called should give jitter in terms of fractional pixels. Ifpixelscaleis the physical dimension of one pixel,the ``scaleparameter provided when Smear is called should give jitter in terms of the same units.oversample (int, optional) – Oversampling factor of img. Default is 1.
- Returns:
out – Image with smear applied.
- Return type:
ndarray
Examples
Apply 25 pixels smear at 30 degrees to a 5x oversampled PSF:
>>> import matplotlib.pyplot as plt >>> import lentil >>> psf = ... # PSF calculation details omitted >>> psf_smear = lentil.smear(psf, distance=25, ... angle=30) >>> plt.subplot(121), plt.imshow(psf) >>> plt.subplot(122), plt.imshow(psf_smear)
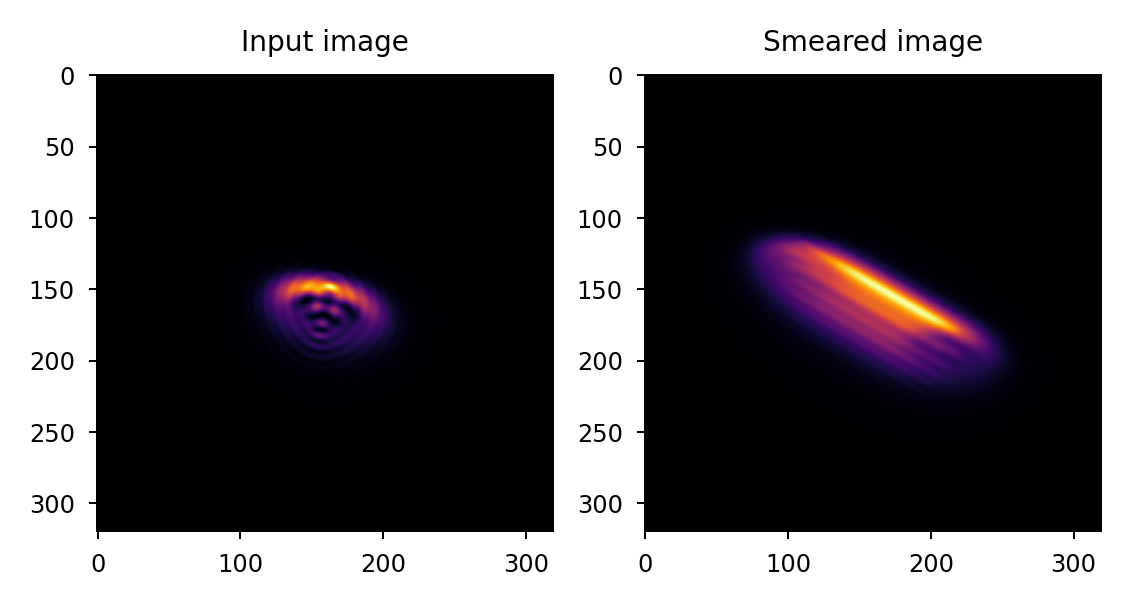
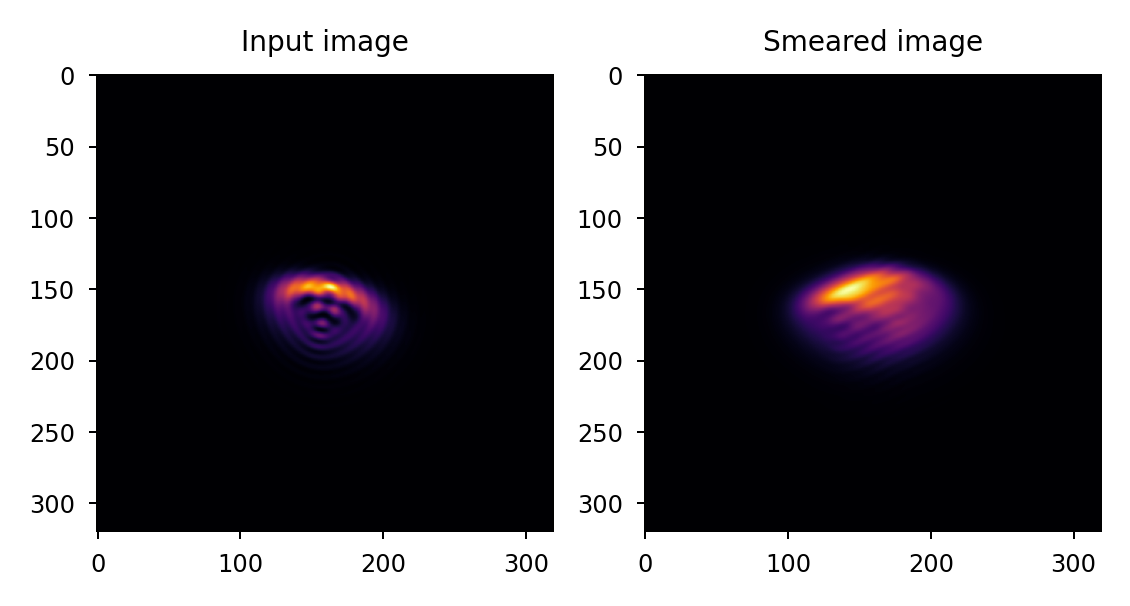
Apply 50 um smear at a random angle to a 5x oversampled PSF. Note that because we are specifying smear in terms of linear distance on the focal plane, we must also provide the detector pixelscale:
>>> import lentil >>> import matplotlib.pyplot as plt >>> psf = ... # PSF calculation details omitted >>> psf_smear = lentil.smear(psf, distance=5e-5, ... pixelscale=5e-6, oversample=5) >>> plt.subplot(121), plt.imshow(psf) >>> plt.subplot(122), plt.imshow(psf_smear)
References